What does it mean to "root" a phylogenetic tree? Trees drawn using simple methods like neighbor-joining (NJ) are unrooted. The tree's horizontal structure reflects "distances" between the sequences being compared, but the choice of which internal node lies to the extreme left (the root) is usually arbitrary. The standard way to root a tree produced by this method is to include an outgroup in the set being analyzed---a sequence for which it is "known" that all the other sequences are more closely related among themselves than to the outgroup. Then, we know that the ancestral sequence lies on the evolutionary path between the outgroup and all the other sequences.
The tree which I showed last time was made by Clustal using the NJ algorithm.
The newlines in this are not relevant (although the formatting helps us to see the structure), so this representation is equally valid:
When I loaded the tree into R
I had this:
:
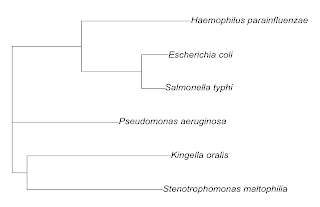
Note that APE considers it to be unrooted. We can confirm the position designations of the tips by the following code. We first do the plot, suppressing the tip labels (species names), then add the tip labels. The first argument to the function tiplabels is the vector of labels, the second is the vector of tip positions to be labeled. We see that they are numbered simply from bottom to top.

And we can find the internal nodes as follows:

We can root the tree by picking the outgroup (node #2 = Kingella oralis) as follows:

If I write the data for the new tree to disk
I get:
which is not quite the same as what we started with. Replacing the species by two letter abbreviations and removing the lengths of the terminal branches, we started with:
Now we have:
The distance (0.00827) which was previously assigned to the SM,KO clade (nodes 1,2) is now assigned to the PA,ST,EC,HP clade (nodes 3-6).